Load libraries
library(tidyverse)
library(lavaan)
library(semTools)Multiple Regression and Beyond (3e) by Timothy Z. Keith
library(tidyverse)
library(lavaan)
library(semTools)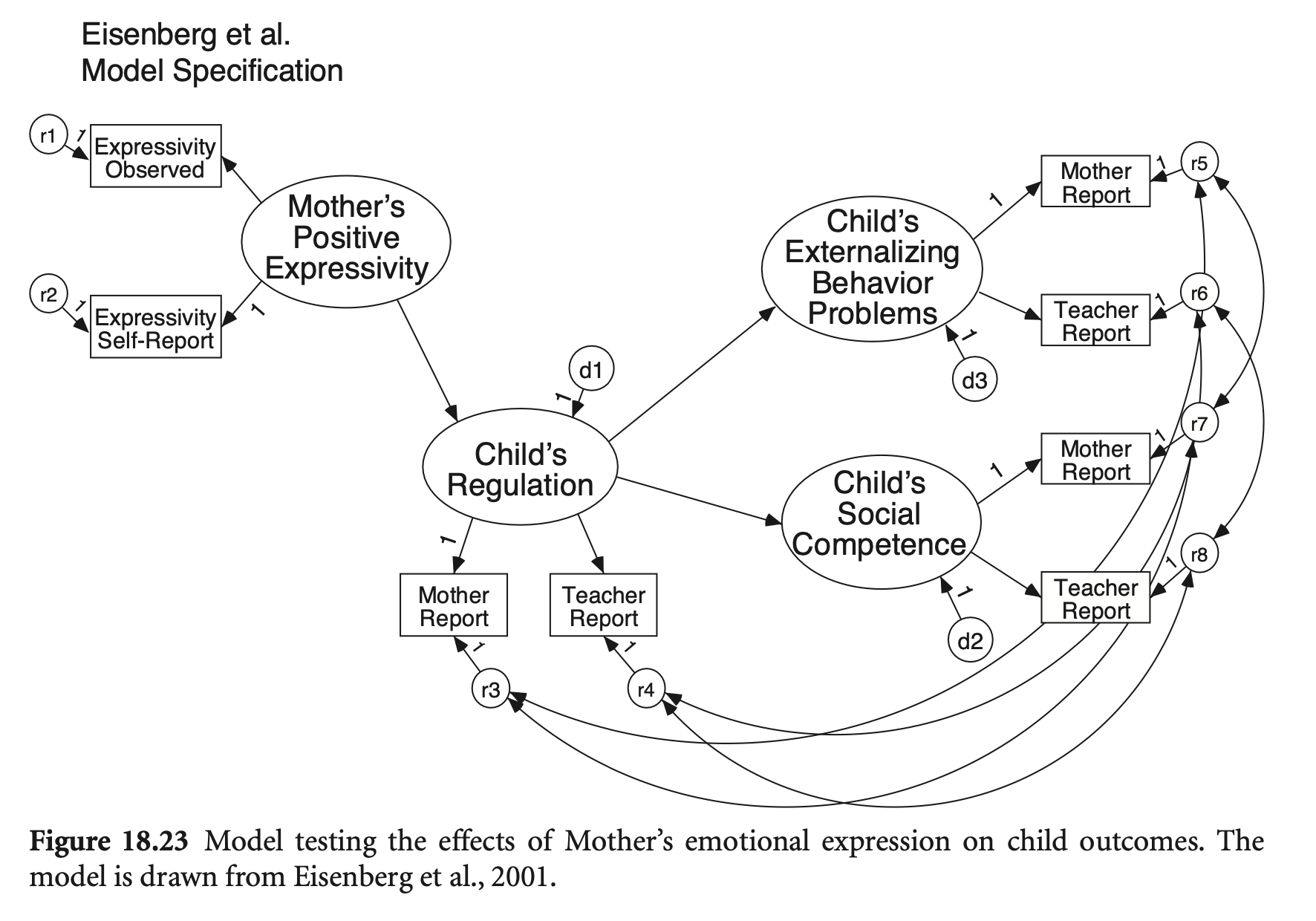
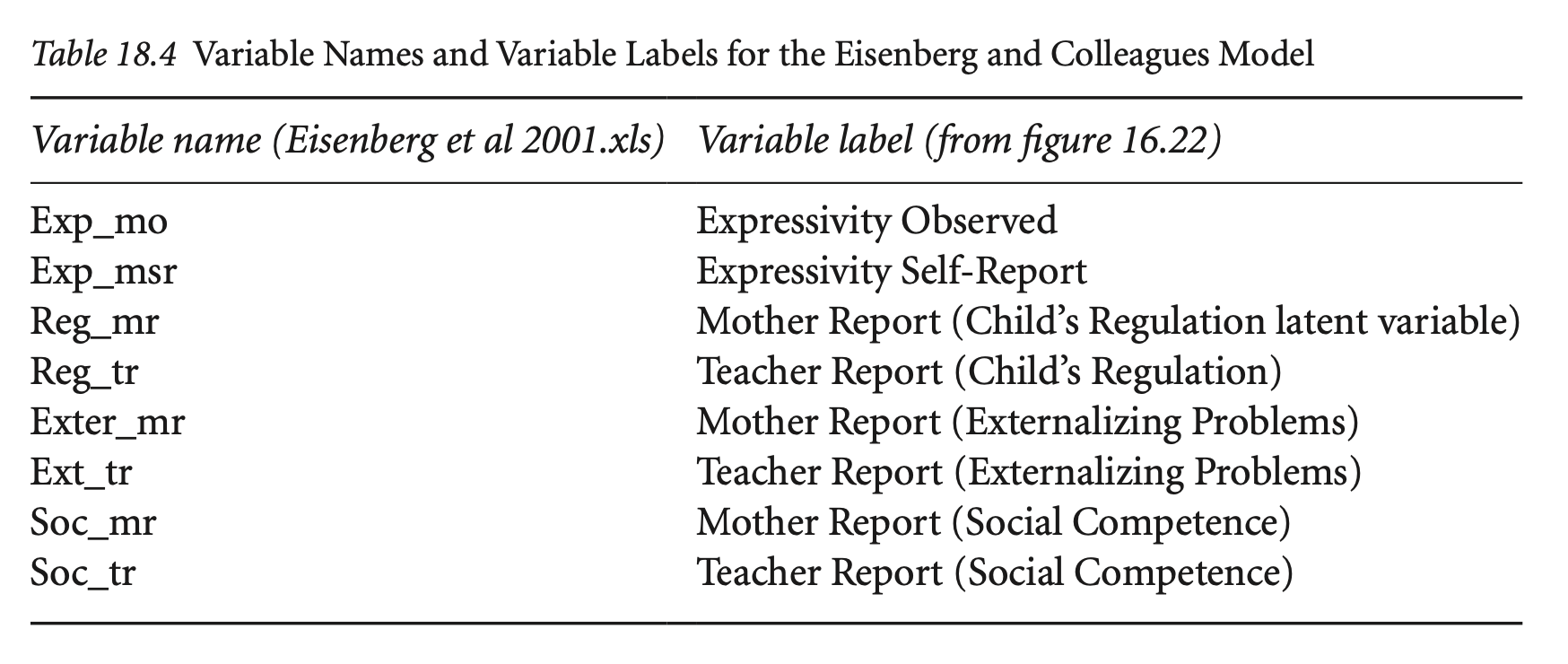
Residual covariance에 대한 제약
# load data
eisenberg <- haven::read_sav("data/chap 18 latent var SEM 2/Eisenberg et al 2001.sav") |>
rename(ext_mr = exter_mr)
eisenberg |> print()# A tibble: 176 × 8
exp_mo exp_msr reg_mr reg_tr ext_mr ext_tr soc_mr soc_tr
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 -1.70 6 -0.257 -0.474 29 9 0.350 0.402
2 2.19 10 1.49 1.51 1 4 0.509 0.706
3 0.959 6 -0.628 -1.50 27 22 0.00530 -1.71
4 0.925 7 1.24 -0.00363 23 15 1.49 -0.825
5 1.66 6 -0.567 0.455 17 14 -0.386 0.299
6 1.62 7 -1.80 -0.530 24 15 -1.60 -0.128
# ℹ 170 more rows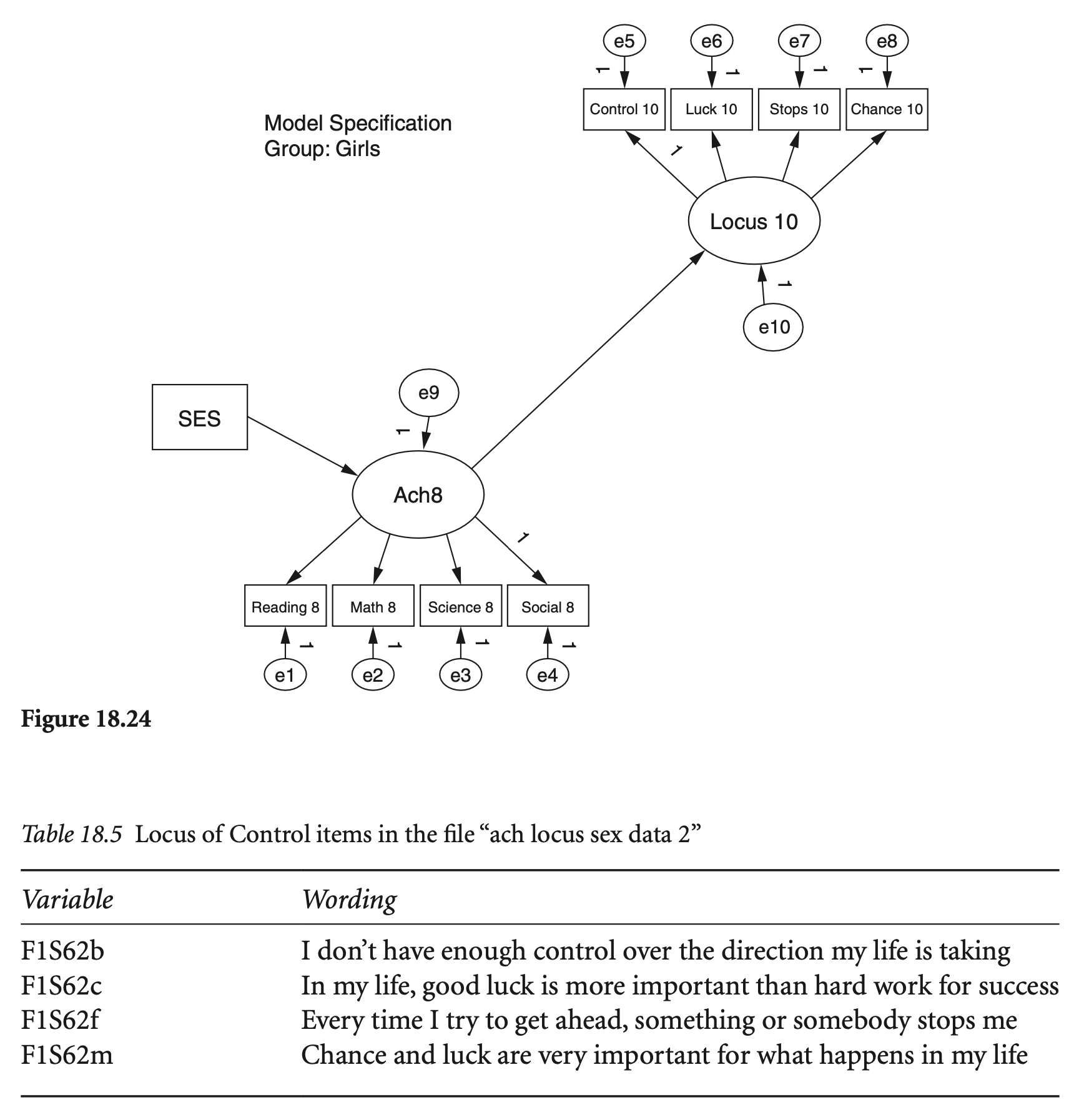
# load data
nels <- haven::read_sav("data/chap 18 latent var SEM 2/ach locus sex data 2.sav")
nels |> print()# A tibble: 857 × 10
byses sex bytxrstd bytxmstd bytxsstd bytxhstd f1s62b f1s62c f1s62f f1s62m
<dbl+lbl> <dbl+lbl> <dbl+lb> <dbl+lb> <dbl+lb> <dbl+lb> <dbl+l> <dbl+l> <dbl+l> <dbl+l>
1 -0.563 2 [Female] 65.5 59.5 64.7 67.8 3 [DIS… 3 [DIS… 3 [DIS… 2 [AGR…
2 0.123 1 [Male] 50.6 48.0 49.1 46.9 2 [AGR… 3 [DIS… 2 [AGR… 2 [AGR…
3 0.229 2 [Female] 59.1 42.8 49.1 47.9 2 [AGR… 2 [AGR… 3 [DIS… 2 [AGR…
4 0.687 1 [Male] 44.4 49.2 35.5 57.3 3 [DIS… 3 [DIS… 3 [DIS… 3 [DIS…
5 0.633 2 [Female] 63.3 52.5 51.8 52.1 2 [AGR… 3 [DIS… 2 [AGR… 3 [DIS…
6 0.992 1 [Male] 31.8 44.5 39.5 38.3 3 [DIS… 3 [DIS… 3 [DIS… 3 [DIS…
# ℹ 851 more rows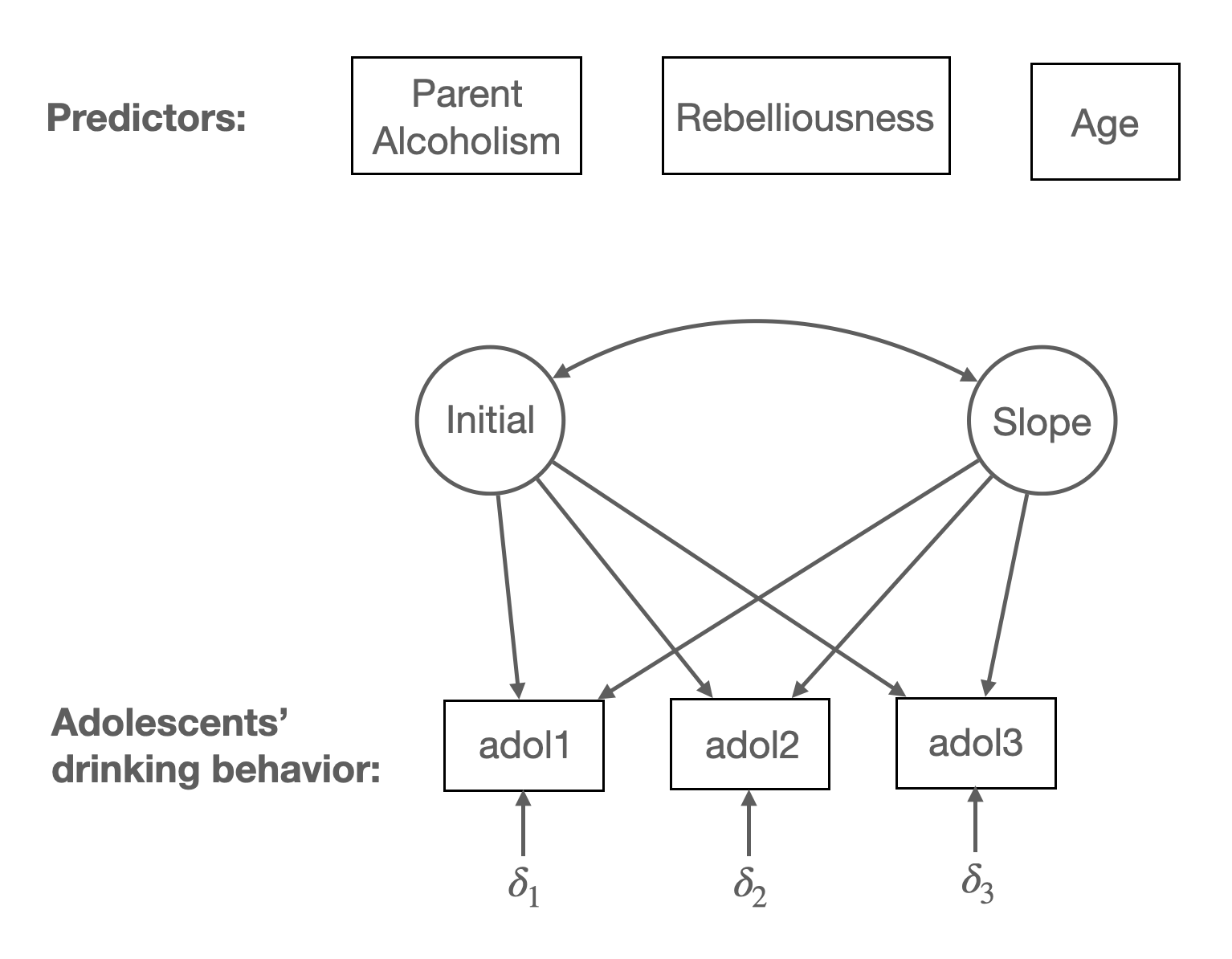
# load data
vars <- c("adol1", "adol2", "adol3", "rebel1", "age", "sex", "parent")
sd <- c(2.81, 3.98, 4.79, 0.69, 1.4, 0.5, 0.49)
lower <- "
1
0.68 1
0.5 0.68 1
0.3 0.29 0.32 1
0.31 0.31 0.23 0.14 1
0.01 0.06 0.1 0.14 -0.03 1
0.12 0.17 0.2 0.12 -0.15 0 1
"
alcohol <- getCov(lower, names = vars, sd = sd)
mean <- c(1.36, 2.12, 3.18, 2.58, 12.91, 0.52, 0.56)데이터가 아닌 공분산과 평균으로 추정하니, 다음과 같이 모형을 추정할 때, 표본의 공분산 행렬, 평균값, 표본 수를 이용해 진행하세요.
growth(model, sample.cov = alcohol, sample.mean = mean, sample.nobs = 363)